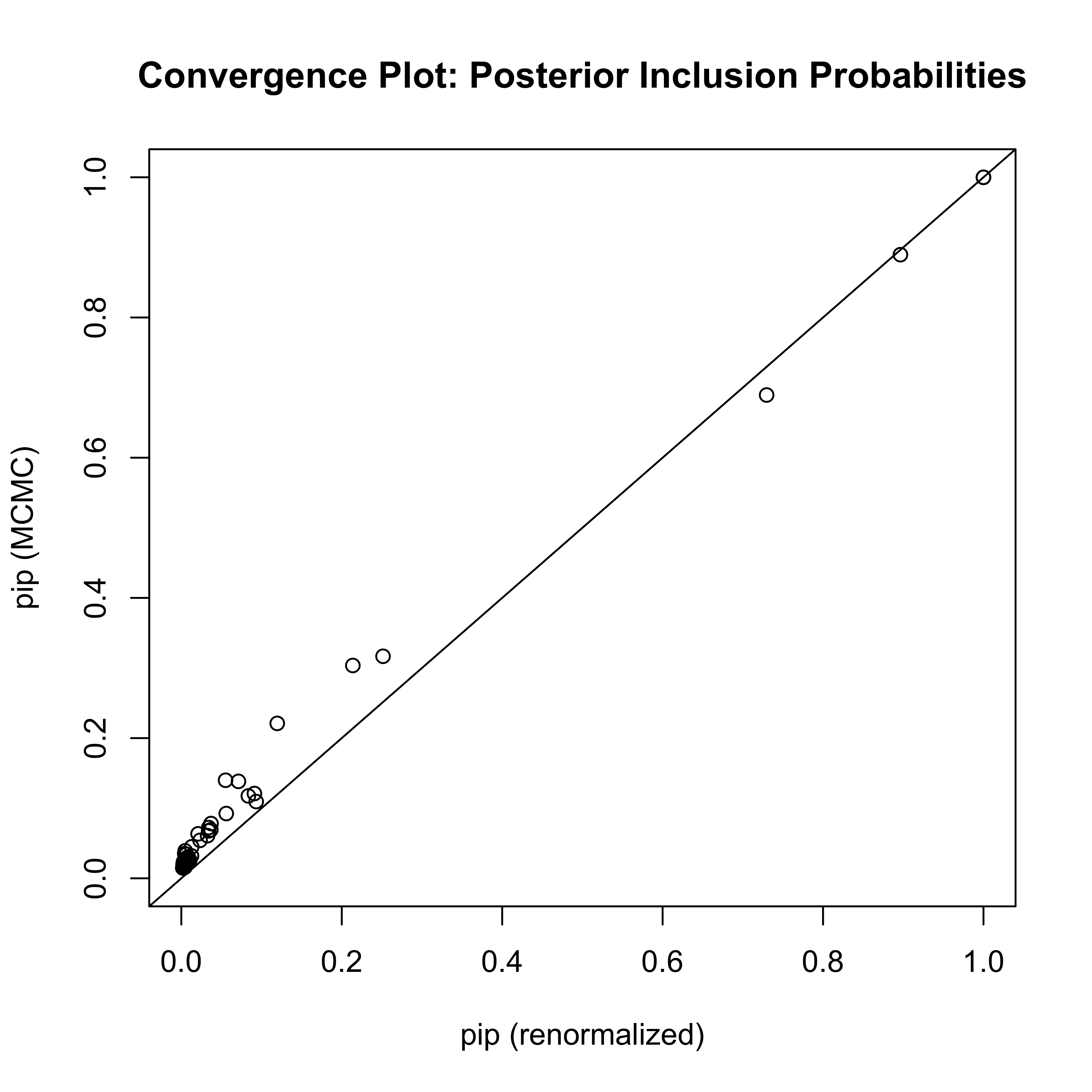
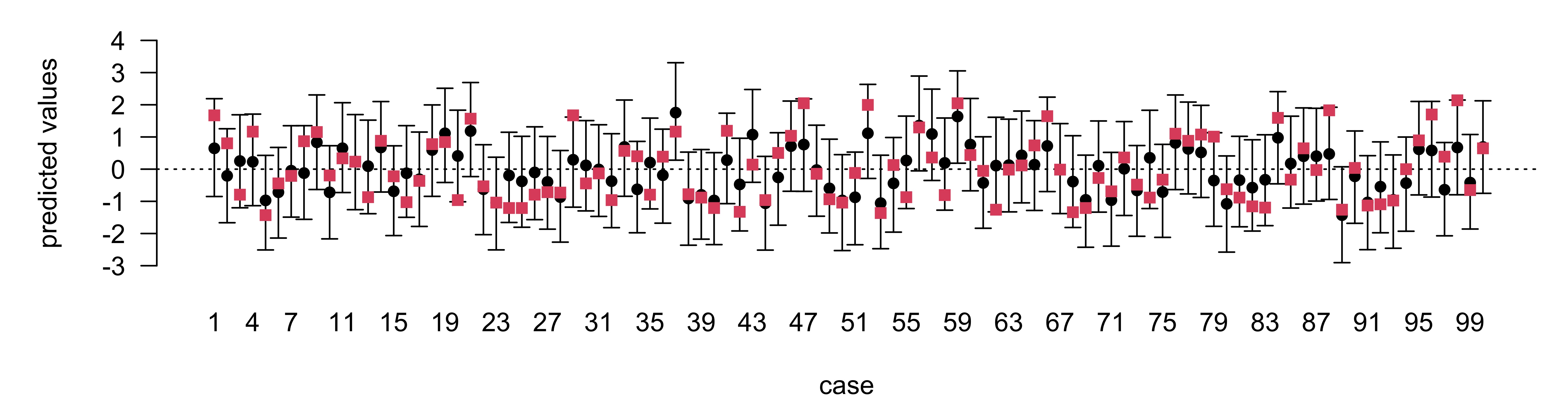
set.seed(8675309)
source("yX.diabetes.train.txt")
diabetes.train = as.data.frame(diabetes.train)
source("yX.diabetes.test.txt")
diabetes.test = as.data.frame(diabetes.test)
colnames(diabetes.test)[1] = "y"
str(diabetes.train)'data.frame': 342 obs. of 65 variables:
$ y : num -0.0147 -1.0005 -0.1444 0.6987 -0.2222 ...
$ age : num 0.7996 -0.0395 1.7913 -1.8703 0.113 ...
$ sex : num 1.064 -0.937 1.064 -0.937 -0.937 ...
$ bmi : num 1.296 -1.081 0.933 -0.243 -0.764 ...
$ map : num 0.459 -0.553 -0.119 -0.77 0.459 ...
$ tc : num -0.9287 -0.1774 -0.9576 0.256 0.0826 ...
$ ldl : num -0.731 -0.402 -0.718 0.525 0.328 ...
$ hdl : num -0.911 1.563 -0.679 -0.757 0.171 ...
$ tch : num -0.0544 -0.8294 -0.0544 0.7205 -0.0544 ...
$ ltg : num 0.4181 -1.4349 0.0601 0.4765 -0.6718 ...
$ glu : num -0.371 -1.936 -0.545 -0.197 -0.979 ...
$ age^2 : num -0.312 -0.867 1.925 2.176 -0.857 ...
$ bmi^2 : num 0.4726 0.1185 -0.0877 -0.6514 -0.2873 ...
$ map^2 : num -0.652 -0.573 -0.815 -0.336 -0.652 ...
$ tc^2 : num -0.091 -0.6497 -0.0543 -0.6268 -0.6663 ...
$ ldl^2 : num -0.289 -0.521 -0.3 -0.45 -0.555 ...
$ hdl^2 : num -0.0973 0.8408 -0.3121 -0.2474 -0.5639 ...
$ tch^2 : num -0.639 -0.199 -0.639 -0.308 -0.639 ...
$ ltg^2 : num -0.605 0.78 -0.731 -0.567 -0.402 ...
$ glu^2 : num -0.578 1.8485 -0.4711 -0.6443 -0.0258 ...
$ age:sex: num 0.69 -0.139 1.765 1.609 -0.284 ...
$ age:bmi: num 0.852 -0.142 1.489 0.271 -0.271 ...
$ age:map: num 0.0349 -0.3346 -0.5862 1.1821 -0.3025 ...
$ age:tc : num -0.978 -0.246 -1.927 -0.72 -0.244 ...
$ age:ldl: num -0.803 -0.203 -1.504 -1.2 -0.182 ...
$ age:hdl: num -0.7247 0.0147 -1.2661 1.6523 0.1046 ...
$ age:tch: num -0.254 -0.176 -0.31 -1.598 -0.216 ...
$ age:ltg: num 0.0644 -0.2142 -0.163 -1.1657 -0.3474 ...
$ age:glu: num -0.636 -0.239 -1.359 0.071 -0.438 ...
$ sex:bmi: num 1.304 0.935 0.915 0.142 0.635 ...
$ sex:map: num 0.258 0.289 -0.381 0.5 -0.697 ...
$ sex:tc : num -1.02 0.131 -1.051 -0.274 -0.112 ...
$ sex:ldl: num -0.927 0.236 -0.913 -0.638 -0.452 ...
$ sex:hdl: num -0.647 -1.188 -0.377 1.189 0.238 ...
$ sex:tch: num -0.411 0.47 -0.411 -1.062 -0.296 ...
$ sex:ltg: num 0.2988 1.2093 -0.0866 -0.6032 0.4857 ...
$ sex:glu: num -0.6171 1.6477 -0.8069 -0.0239 0.7283 ...
$ bmi:map: num 0.189 0.191 -0.477 -0.195 -0.702 ...
$ bmi:tc : num -1.5061 -0.0595 -1.1853 -0.3231 -0.3239 ...
$ bmi:ldl: num -1.267 0.183 -0.976 -0.407 -0.536 ...
$ bmi:hdl: num -0.869 -1.41 -0.286 0.586 0.251 ...
$ bmi:tch: num -0.505 0.505 -0.484 -0.614 -0.388 ...
$ bmi:ltg: num 0.1014 1.1613 -0.4085 -0.5893 0.0716 ...
$ bmi:glu: num -0.862 1.693 -0.89 -0.337 0.358 ...
$ map:tc : num -0.687 -0.148 -0.131 -0.451 -0.21 ...
$ map:ldl: num -0.5407 0.0388 -0.1034 -0.6114 -0.036 ...
$ map:hdl: num -0.235 -0.672 0.254 0.745 0.252 ...
$ map:tch: num -0.29 0.207 -0.258 -0.835 -0.29 ...
$ map:ltg: num -0.214 0.428 -0.427 -0.811 -0.748 ...
$ map:glu: num -0.541 0.659 -0.314 -0.23 -0.812 ...
$ tc:ldl : num -0.144 -0.551 -0.139 -0.509 -0.581 ...
$ tc:hdl : num 0.8363 -0.3457 0.6304 -0.2579 -0.0392 ...
$ tc:tch : num -0.405 -0.326 -0.404 -0.295 -0.451 ...
$ tc:ltg : num -0.901 -0.259 -0.571 -0.392 -0.569 ...
$ tc:glu : num 0.0202 0.0196 0.2073 -0.396 -0.4283 ...
$ ldl:hdl: num 0.889 -0.446 0.705 -0.207 0.26 ...
$ ldl:tch: num -0.463 -0.243 -0.463 -0.21 -0.506 ...
$ ldl:ltg: num -0.6536 0.2724 -0.3783 -0.0708 -0.5638 ...
$ ldl:glu: num -0.0194 0.4995 0.1032 -0.4013 -0.6234 ...
$ hdl:tch: num 0.703 -0.5 0.692 0.171 0.651 ...
$ hdl:ltg: num 0.0179 -1.9846 0.3839 0.0399 0.3043 ...
$ hdl:glu: num 0.654 -2.948 0.689 0.452 0.113 ...
$ tch:ltg: num -0.592 0.531 -0.574 -0.253 -0.537 ...
$ tch:glu: num -0.371 1.114 -0.362 -0.522 -0.34 ...
$ ltg:glu: num -0.584 2.184 -0.468 -0.526 0.183 ...